
Mark C. Fishman, M.D.
- Professor of Stem Cell and Regenerative Biology
- Principal Faculty
Harvard Stem Cell Institute - Board of Directors
Beam Therapeutics - Co-Founder and Chair of the Scientific Advisory Board
Aditum Bio
Mark C. Fishman’s laboratory seeks (1) to unravel the heart-brain axis, using the larval zebrafish to define the circuitry and function of autonomic control of cardiac function, and (2) to understand the genetic and neuronal structure of social behavior in vertebrates.
In the 1990s, by harnessing the first large-scale genetic screens in zebrafish (performed in collaboration with W. Driever and contemporaneously with C. Nuesslein-Volhard), and by providing much of the early genomic infrastructure, Fishman’s lab helped to make the zebrafish a cornerstone of developmental biology, and led to revelation of many of the pathways that guide vertebrate organ development, particularly the heart and vessels.
From 2002 to 2016, Fishman was the founding President of the Novartis Institutes for BioMedical Research (NIBR). During his tenure, NIBR discovered and brought through successful clinical trials 90 new medicines in more than 120 indications. Fishman brought a particular focus on regenerative medicines as treatments for disorders of aging. He has continued his interest in therapeutics, with Professor Melton inaugurating a new Harvard Masters program in Biotechnology of Life Sciences, combined with a Harvard MBA. He serves on the Board of Directors of Beam Therapeutics, and is a founder and SAB Chair of Aditum Bio.
Fishman graduated from Yale College and Harvard Medical School, and was a resident and Chief Resident in Medicine, and fellow in Cardiology, at the MGH and later Chief of the Cardiology Division and Director of the Cardiovascular Research Center at Harvard Medical School and MGH. In addition to his publications in developmental biology and drug discovery, Fishman is the author of the medical textbook, Medicine, and of the book Lab: Building a Home for Scientists, on the history and architectural design of buildings for scientists. Fishman is a member of the National Academy of Medicine, where he recently served two terms on the Executive Committee and Council, and is a Fellow of the American Academy of Arts and Sciences.
Lab Overview
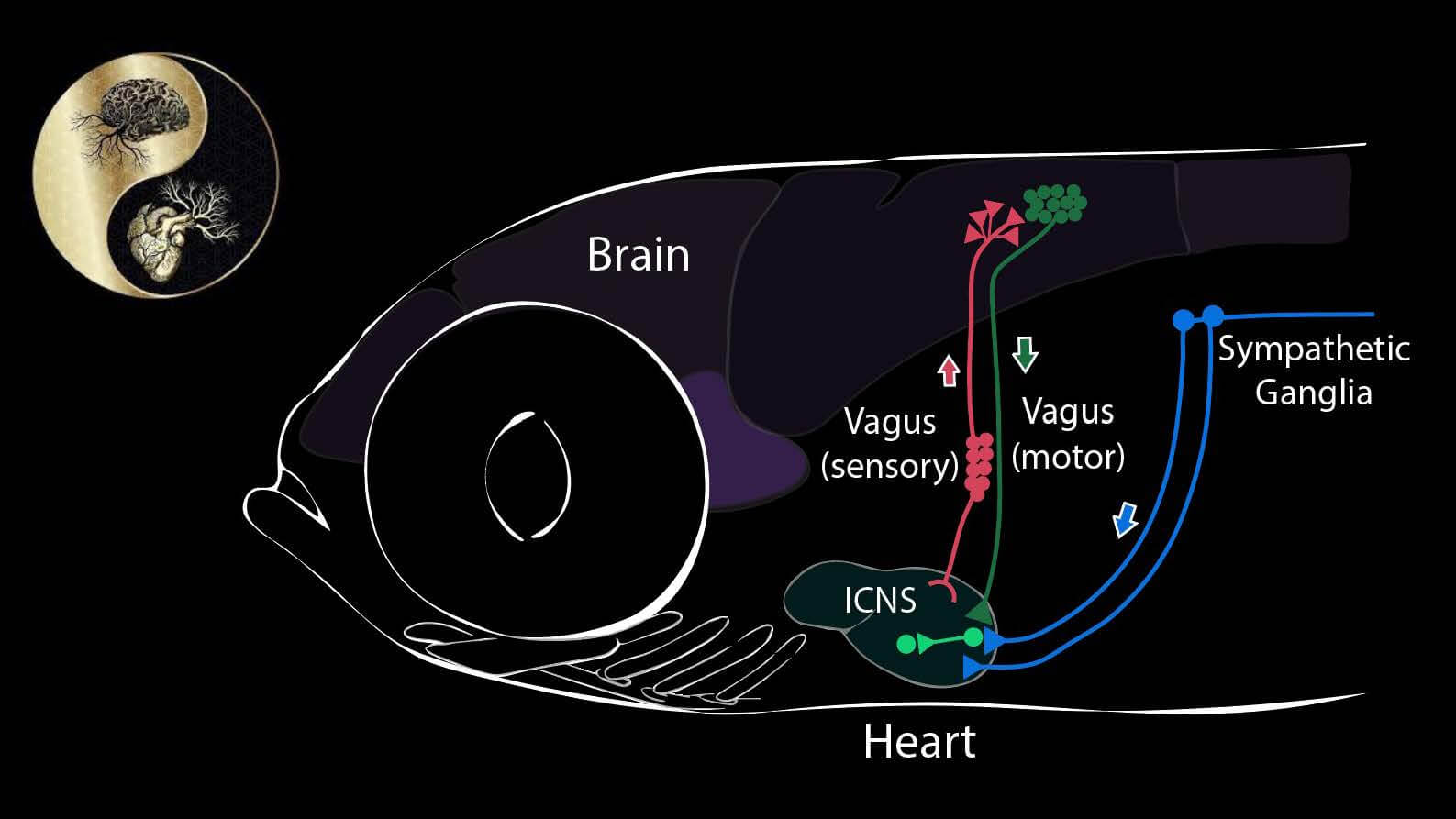
What the heart tells the brain (and vice versa)
Our goal in this project is to discover signals that flow from internal organs to the brain – what parameters are sensed, by what cells, and to what end, in terms of maintaining constancy of the internal environment. We focus in particular upon the heart and cardiovascular system, using the transparent larvae of the zebrafish. The heart, like all internal organs, has its own local mini-brain. This local network is connected to the autonomic nervous system, known to register external threats and internal challenges, such as hypotension and hypoxia. We have found that there are sensory nerve cells (so-called “interoceptors”) that sense other chemical and physical attributes as well. By single cell transcriptional profiling we have built an atlas of cells responsible both for sensing input from the cardiovascular system and for driving the cardiovascular responses. We test these models using cell-directed optogenetic activation or inhibition.
There are several medical implications to this work. When the system falters, as in the common disorders termed “dysautonomias”, the consequences for patients are dire, including syncope and arrhythmias. These problems can arise after viral illnesses, including in long Covid. In addition, there is growing evidence that input from visceral organs affects mood and emotional responses, and hence psychiatric state.
The work is done in collaboration with the labs of Florian Engert (in MCB) and Misha Ahrens (at Janelia Farms). Projects range across a host of technologies, including single cell transcriptional analysis, in vivo neuronal activity monitoring, transgenesis, optogenetics, patch clamp analysis, electron microscopy and expansion microscopy.
Genetics of Collective Behavior
Our goal in this project is to discover the genetic, cellular, and neural pathways that govern social behaviors.
Animals live in conspecific groups, as schools of fish, flocks of birds, or tribes of humans. This behavior provides evolutionary advantages in foraging for food, avoidance of predators, and choice of migratory routes, but also requires sacrifices on the part of individuals. What are the forces that direct individuals to participate in and lead such groups? How are they communicated? Are these behaviors encoded In the nervous system and how are they translated into action? How and when do these behaviors develop?
The medical implications of this work are profound. Many psychiatric disorders, including autism and schizophrenia, are manifest primarily as deficits in certain social abilities. Are the genes associated with psychiatric illness among those dedicated to social interactions? Are their roles conserved from fish to man?
In collaboration with Florian Engert’s lab, we are using zebrafish to understand how genetic changes in individuals affect how groups form and behave. Collective behavior is an “emergent” group property. We have discovered that mutations in in zebrafish genes related to human disorders of social behavior, such as autism or schizophrenia, change how the adult fish swim in groups, for example, tending to scatter or huddle more than do wild-type. These same tendencies are evident even in young larvae. Simple algorithms, based on how an individual larval fish responds to dots moving in its environment, can predict how the fish behave in groups, and can account for a large proportion of the differences in the mutant fish. This gives us the chance to understand, at a single cell level, what neural changes can account for the different responses. Because we have found that drugs useful in patients with psychiatric disorders can reverse certain of the behavioral differences in mutant fish, we can also use this system to screen for novel therapeutics.
